Co-Infection Dynamics of Influenza A Virus and Defective Interfering Particles
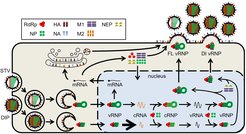
Motivation
Defective interfering particles (DIPs) are homologous to their corresponding standard virus (STV) particle, contain at least one defective genome segment, and can strongly inhibit the production of infectious virus particles. Infection experiments have shown that the application of DIPs can rescue mice from lethal doses of influenza A virus (IAV) [1]. Furthermore, IAV-derived DIPs can enhance the innate immune response in a host and induce antiviral activity against various viruses, e.g., RSV and SARS-CoV2. Thus, DIP preparations are considered for antiviral therapy. While DIP biology is relatively well understood, many quantitative aspects of DIP de novo generation, replication and interference remain largely unknown. To further the development of DIPs as antivirals, a more comprehensive understanding of the underlying mechanisms of DIP activity is required.
Aim of the project
Based on previously established IAV infection models [2], we develop mathematical models covering the virus dynamics on the intracellular and cell population level during STV and DIP co-infection [3-5]. For model calibration, we employ quantitative experimental data obtained via state-of-the-art analytical methods to describe virus dynamics for different co-infection scenarios. Based on our models, we can better predict infection dynamics and characterize virus spreading to identify optimal DIP dosing for antiviral treatment. Furthermore, the models support the optimization of influenza vaccine production processes and the development of therapeutic approaches against a wide range of other viral infections.
