Multiscale modeling of influenza A virus infection in highly different infection conditions
Motivation
The mechanisms of intracellular influenza virus replication and virus spreading involve highly complex processes. This concerns, for example, the copying of the viral genome inside infected cells, an adaptive viral defense against cellular immune responses, a multi-layered regulation of these processes by viral proteins or the spread of virus particles between neighboring cells and in cell populations. The spatial and temporal dynamics of such an infection are strongly influenced by the amount of infective virus particles, i.e., the multiplicity of infection (MOI). High MOIs result in faster infections that are more likely to overcome the immune system; low MOIs can result in significantly higher virus titers for cell culture-based vaccine production [5]. During the initial phases of a natural infection, e.g. via airborne transmission of virus particles, mostly low MOIs are relevant [6]. However, during later phases of infection and for virus research, cells are usually infected by a large number of virus particles. Both scenarios need to be considered to fully understand influenza virus infection dynamics.
Defective interfering particles (DIP) are homologous to a standard virus (STV) particles, contain at least one defective genome segment and can severely inhibit the production of infectious virus particles. Thus, they are considered as potential candidates for antiviral therapy. The interaction of STVs and DIPs during infection is highly dependent on the applied virus concentrations, i.e. the MOI and the multiplicity of DIPs (MODIP). Mouse experiments have shown that the application of large amounts of DIPs can completely protect mice from infection-induced death, while lower amounts of DIPs showed weaker or no effects [2,6].
![Influenza A virus infection dynamics: A) Dynamics of cell infection and changes in the MOI over the time course of an animal cell culture infection. B) Percentage of infected cells for low, medium and high MOIs [3].](/4207580/original-1705048343.jpg?t=eyJ3aWR0aCI6MjQ2LCJvYmpfaWQiOjQyMDc1ODB9--a8196609bfbb7f265b88e4027d8e11b0c5a6b49a)
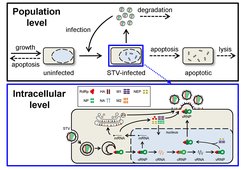
Aim of the project
Novel experimental methods enable in-depth investigations of influenza virus infections on the intracellular and the cell population level [4]. Based on a model previously established in our group [7], we aim to develop an MOI-sensitive multiscale model that describes virus replication and propagation dynamics in highly different conditions using a single set of model parameters. Additionally, we want to incorporate the interaction of STVs and DIPs during infection to evaluate how the MOI and MODIP affect infection dynamics[1]. Such models enable the prediction of infection dynamics for a large range of infection conditions. Furthermore, they can be used to support the optimization of vaccine production processes and the development of therapeutic approaches against influenza.

![Influenza A virus infection dynamics: A) Dynamics of cell infection and changes in the MOI over the time course of an animal cell culture infection. B) Percentage of infected cells for low, medium and high MOIs [3]. Influenza A virus infection dynamics: A) Dynamics of cell infection and changes in the MOI over the time course of an animal cell culture infection. B) Percentage of infected cells for low, medium and high MOIs [3].](/4207580/original-1705048343.jpg?t=eyJkb19ub3RfdHJhbnNmb3JtIjp0cnVlLCJmaWxlX2V4dGVuc2lvbiI6ImpwZyIsIm9ial9pZCI6NDIwNzU4MH0%3D--63309bb73ecef066df0362c7b2dc92bfb758a101)