Modeling the Dynamics of Influenza Virus Replication in Mammalian Cells
Motivation
Influenza viruses hijack a host cell's biosynthetic machinery to produce viral proteins and replicate their genome. This process has to be highly regulated to yield large amounts of progeny virus particles despite the antiviral mechanisms that cells have evolved. Over the last decades substantial progress has been made towards understanding the molecular mechanisms that mediate influenza replication. However, most of this knowledge is only qualitative and much remains to be learned about quantitative aspects and the dynamics of virus-host cell interaction. Here, we combine concepts from systems and molecular biology to establish dynamic models of influenza virus replication in mammalian cells. Mathematical analysis and experimental validation form an iterative cycle, which allows refining our models based on the increasing biological knowledge.
Aim of the project
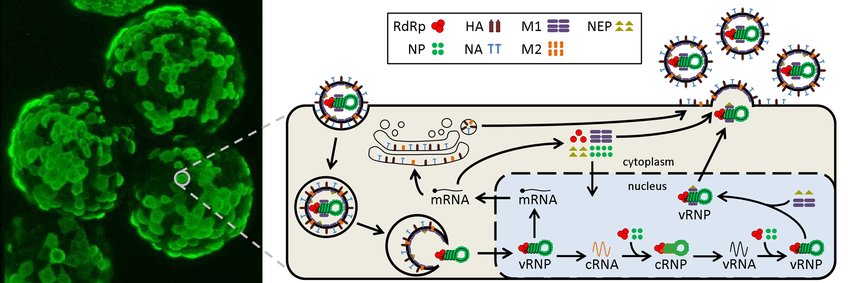
With respect to state-of-the-art vaccine production and models already developed by our group [1-5], we investigate influenza virus replication in MDCK cells. Within each cell, the virus synthesizes three viral RNA species and up to eighteen viral proteins. We model these components quantitatively to determine their influence on virus infectivity and yield. In addition, we consider modeling of an individual cell is a crucial step towards understanding the infection dynamics in cell populations.
In collaboration with the Molecular Biology group various experimental methods are used to validate the resulting model predictions in order to:
- understand the regulation of virus replication in mammalian cells,
- determine the bottleneck for virus yield,
- elucidate virus-host cell interactions,
- understand the impact of defective interfering virus particles on virus propagation, and
- develop novel optimization strategies for vaccine production.