Methods and Tools for Metabolic Modeling and Design
We develop various mathematical approaches for analyzing molecular networks in the cell. One particular focus is theoretical and computational methods for the analysis and targeted modification of metabolic networks based on stoichiometric and constraint-based modeling approaches. Relevant aspect of our research include:
- Analysis of solution spaces arising in stoichiometric modeling and flux balance analysis of metabolic networks.
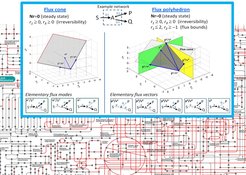
- Theory of elementary flux modes and of elementary flux vectors for metabolic pathway analysis.
- Computational strain design and theory of minimal cut sets for targeted (re)design of metabolic networks.
- Theory of growth-coupled product synthesis.
- Efficient inclusion of enzyme (and other) constraints in constraint-based metabolic models.
- Methods for reduction of genome-scale metabolic networks to core models.
- Analysis of thermodynamic driving forces in metabolic networks and the role of the redundant redox cofactors NADH/NADPH in the cellular metabolism.
- Constraint-based modeling and design of microbial communities (e.g., communities involved in anaerobic digestion).
Previously, we have also been developing various tools and methods for qualitative and semi-quantitative modeling of signaling and regulatory networks based on interaction graphs, logical networks, and logic-based ODEs and used them to analyze large-scale mammalian signaling networks.
Software packages
We have been developing several software packages for the analysis of metabolic (and other biomolecular) networks, which also integrate many of our theoretical developments. The most important packages are listed here and further toolboxes can be found on our group’s GitHub repositories.
The two largest toolboxes are CellNetAnalyzer (CNA; MATLAB/Octave-based) and CNApy (CellNetAnalyzer for Python), each providing a graphical front-end for the intuitive analysis of metabolic networks with various standard and advanced constraint-based methods. CNA supports in addition the analysis of signal transduction and (gene) regulatory networks based on logical networks and interaction graphs as modeling formalisms. Both CNA and CNApy are widely used in the scientific community, also for teaching. CNA and CNApy are resources of the German bioinformatics infrastructure network de.NBI, within which we and other groups offer extended user support and training workshops for our tools.


