Using Functional Metagenomics to Discover Novel Enzymes for Glycan Analysis
Most current methods to detect glycans and resolve their structures require enzymatic treatment coupled to analytical techniques. Enzymes termed endoglycosidases that release intact glycans from glycoconjugates are used at the front end of workflows that use “free” glycans as starting material. Highly specific exoglycosidases are often used to sequentially remove sugar units one by one to enable elucidation of a glycan’s composition. Some glycans can be modified with additional chemical groups (e.g., sulfate, phosphate etc) in specific positions, often called post-glycosylation modifications (PGMs). While already a bunch of exo- and endo-glycosidases is available for use in analytical workflows, some enzymatic activities are still missing in the glycoanalytical toolbox to deconvolute the enormous structural complexity of the glycome (Figure 1). Additionally, enzymes (e.g. sulfatases) that can remove specific PGM’s to enable precise glycan structural analyses are not yet established. To facilitate discovery of missing enzymatic tools, we developed a functional metagenomics workflow to enable discovery of novel enzymes encoded by environmental DNA.

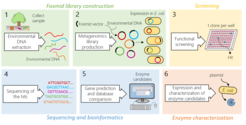
Microorganisms are the most abundant organisms found in nature. They have adapted to live in any given environment ranging from the human gut to hot springs resulting in tremendous bio- and genetic diversity. The pool of microbes populating earth represents a goldmine for new enzyme discovery. Yet, accessing this goldmine remains challenging due to the very small portion (~1-2%) of microorganisms that can be successfully cultivated in laboratories. To circumvent this limitation, the technique of functional metagenomics uses direct extraction of DNA from all microorganisms living in a specific environment without any cultivation (termed eDNA or environmental DNA). This facilitates access to genetic information from both cultivable and uncultivable microorganisms. In the process of functional metagenomics, isolated eDNA is introduced into a laboratory host to permit micro-expression of many environmental genes. In our workflow, isolated DNA is cloned into a fosmid vector prior to introduction into the laboratory host Escherichia coli. Resulting metagenomics libraries are arrayed in 384 well plates in which each well contains a unique E. coli clone harboring a different piece of environmental DNA (~40 kb). These libraries are then screened using reporter substrates, typically fluorescent monosaccharides, to seek new glycoside hydrolases with desired activities. Pieces of environmental DNA from the obtained hits are then sequenced to identify the gene responsible for the observed activity. Finally, interesting enzyme candidates are tested for their ability to function within glycoanalytical workflows (Figure 2). In this project, screening has already yielded identification of both new glycoanalytical tools and novel enzyme families.
Collaboration
This project is co-supervised by Dr. Christopher H. Taron at New England Biolabs, Ipswich (USA).

