Metabolic Profiling of Mammalian Cell Culture Systems
Motivation
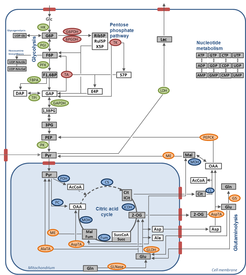
Mammalian cell culture has grown to one of the major production systems for biopharmaceuticals over the last decades. Despite this huge interest of both industry and academia, relatively little is known about the overall regulation linking cell metabolism, cell growth, and productivity. In many cases, modeling approaches like MFA or DOE relying on steady state assumptions are used to describe and predict specific limitations that could decrease cell growth or productivity. A much deeper understanding of cell metabolism can be achieved by dynamic measurements of intracellular metabolite pools, which describe changes in the physiological state of cells over the time course of cultivations. This knowledge is a valuable research tool to understand the function of metabolic pathways within the metabolic network, to investigate changes in energy and precursor pools, and to identify possible bottlenecks for growth and product formation. Using this information, a rational optimization strategy for the improvement of product yields (metabolic engineering) can be performed.
Aim of the project
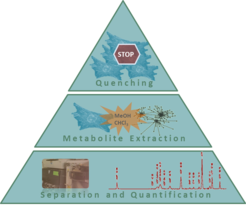
Highly developed sample preparation methods are crucial for an accurate measurement of intracellular metabolites. In particular, the shutdown of cell metabolism (quenching) and the subsequent isolation of metabolites from the cell extract (extraction) need to be optimized to obtain a detailed snapshot of metabolite concentrations. In a next step, for simultaneous quantification of specific metabolic targets, the extracted metabolites are separated due to their charge and size employing anion exchange chromatography. Compounds are detected via conductivity, UV and MS and matched with a calibration standard to finally calculate compound concentrations.
Metabolic response to viral infection
Viral infection of mammalian cells can have a significant effect on various metabolic pathways leading to an alteration in cell growth and morphology. Changes in metabolic pathways of infected cells can lead to increased substrate consumption and by-product formation. Metabolic profiling can help to locate intracellular changes in the metabolic network and identify possible metabolic bottlenecks. Appropriate media supplementation or feeding strategies could then be applied to avoid nutrition limitation during virus production.