Self-Organizing Systems from Functional Biomimetic Modules
Introduction
This project is a part of the MaxSynBio initiative and supports the partners' experimental activities by approaches based on mathematical models. The system wide modeling activities pursue two main goals: On the one hand, behavioral models of artificial cells contribute to the understanding of self-reproducing entities and help to find conditions for auto-poietic behavior. On the other hand, the project considers more technological aspects. The long-term goal is to develop a design methodology for integrated artificial biosystems to be used as biochemical microreactors. The project first provides models of the functional modules within cell-like entities. It uses experimental data and, where available, modeling approaches from the MaxSynBio partners. Based on this information, it generates model formulations of functional units that are applicable in the context of an artificial whole cell model. It feeds back the functional units' models into the experimental working packages. The validated models of the functional units are integrated into a model of a complete artificial cell-like entity. The whole cell models will allow predicting some of the properties of integrated biosystems. Methods of nonlinear system analysis help to understand the interactions between the functional units as well as their synchronization within a cell-like entity. The results of the system analysis lay the ground for systematic construction of integrated biosystems. Based on a model library of functional modules, the project will design artificial cell like entities in silico, which have certain desired properties, e.g. the ability to reproduce or to synthesize a biochemical product.
Results
To model an in-silico protocell from subsystems, models of experimentally validated parts were combined to a fictitious cell-like entity. The objective was to generate a system able to grow and to divide after having doubled its length by combining functional modules for a growing membrane, a positioning module that determines where the system should be divided and a division module. The simulation studies indicate that the constructed protocell behaves in principle as desired [1, 2]. However, they also show that it is not sufficient just to combine parts together, but further coordinating units are necessary to get a robust growth and dividing artificial cell.
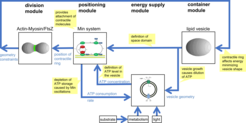
System theoretic view on a cell-like entity consisting of functional modules for containment, energy supply, positioning of a contractile ring, and division. Blue arrows indicate information exchange between functional modules.
Alternatively, we formulate models of synthetic biological devices, so-called functional modules, studied by our partners in experiments. Here, the modeling of functional modules concentrated on the actin-myosin system as part of the division module and the electron transport chain ETC as part of the energy supply module in close cooperation with and the group of Prof Schwille (MPI for Biochemistry, Martinsried) and the PSE group. A comparatively simple continuous actin-myosin model was developed to qualitatively explain the experimental findings by extending existing models. As a novel approach, the dependence of the actin-myosin interaction on the ATP concentration was combined with spatially distributed mass and momentum balances of the involved species. The simulation results [3] show that the model is able to cluster an initially spatially homogeneous F-actin density to one main cluster or to a variety of clusters, according to experimental observations, because of a new formulation for the active contractile stress, which models the dependency of cluster formation on a medium ATP concentration. Recently, a further extension of the subsystem exhibits cortical actin cluster movement similar to in-vitro observations with synthetic minimal actin cortices and helps to understand the underlying cluster migration mechanism [4].
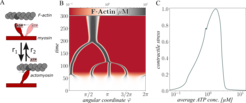
F-actin cluster formation depending on a medium ATP-concentration. A: Underlying simplified myosin cross-bridge model. B: Clustering of distributed F-actin cortex in a 1D ring system. C: Average contractile stress against ATP concentration.
Further, suitable models for the dynamics of the ETC containing systems were developed and fitted to experimental data obtained by the PSE group. In particular, a minimal kinetic model was derived of a proto-metabolism coupled to the ETC containing inverted membrane vesicles (IMVs) from E. coli in droplets. The model includes continuous NAD+ regeneration by the substrate glucose-6-phosphate oxidation [5]. Additionally, a model of a synthetic ETC was developed. In this model, the functionalities of the ETC membrane proteins are mimicked by membrane-situated TCNQ which oxidizes NADH and transfers electrons through a vesicle membrane [6]. The mathematical simulations indicate that the minimal models are able to quantitatively reproduce the experimental findings with a predictive potential, especially in terms of the oxygen consumption of the ETC.
Future Directions
The future work lies on the modeling and experimental validation of further biomimetic functional modules like energy supply, membrane container or positioner module towards a computer-based model library for the in-silico design of protocells.
Cooperation Partners
- PSE group
- group led by Prof. Petra Schwille (MPI for Biochemistry, Martinsried)
- members of the MaxSynBio research network
Publications
[1] Schneider E, Schweizer J, Mangold M. Bringing the parts together: Steps towards an in-silico protocell. IFAC-PapersOnLine. Elsevier B.V.; 2016;49: 20–25.
[2] Schneider E, Mangold M. Modular assembling process of an in-silico protocell. Biosystems. Elsevier Ireland Ltd; 2018;165: 8–21.
[3] Wölfer C, Vogel SK, Mangold M. A curvilinear Model Approach: Actin Cortex Clustering Due to ATP-induced Myosin Pulls. IFAC-PapersOnLine. Elsevier; 2016;49: 103–108
[4] Vogel S, Woelfer C, Ramirez DA, Flassig RJ, Sundmacher K, Schwille P. Cortical Actomyosin Flows are Generated by Actomyosin Cluster Vibrations in Vitro. bioRxiv. 2018; Available: http://biorxiv.org/content/early/2018/08/18/394700.abstract
[5] Wang M, Wölfer C, Otrin L, Ivanov I, Vidaković-Koch T, Sundmacher K. Transmembrane NADH Oxidation with Tetracyanoquinodimethane. Langmuir. 2018;34: 5435–5443.
[6] Beneyton T, Krafft D, Bednarz C, Kleineberg C, Woelfer C, Ivanov I, et al. Out-of-equilibrium microcompartments for the bottom-up integration of metabolic functions. Nature Communications 2018;9: 2391.

