Population Balance Modeling of Virus Replication during Vaccine Production
Motivation/Objectives
Vaccination constitutes the most effective method to prevent contagion with infectious diseases like hepatitis, measles, and influenza. Up to date the majority of infuenza vaccines is produced in embryonated chicken eggs. This process is well-established and relatively cheap, but it also has several disadvantages which can be overcome by cell culture-based production processes. These are already widely-used for other viruses and become increasingly relevant for influenza vaccines as well.
Cell cultures in vaccine production processes show significant cell-to-cell variability in terms of physiological states, which can be quantified experimentally via flow cytometry. Characteristic fluorescence distributions with respect to viral nucleoprotein NP are shown in Figure 1. Those distributions show interesting patterns of behavior including transient multimodality and a backshift to lower fluorescence intensities at later times.
In cooperation with the BPE group our work aims at developing distributed models of the process using Population Balance Modelling to reproduce and predict distribution dynamics of infected cells during virus replication. In contrast to conventional approaches, which focus only on the mean of distributions, this approach explicitly accounts for cell-to-cell variability in order to gain a deeper understanding of the underlying biological processes.

Approach/Results
Compared to our earlier work in this field [1] focus has shifted from one-dimensional unstructured population balance models that are able to mechanistically describe the observed distribution patterns to multi-dimensional population balance equations. These allow incorporation of structured intracellular viral replication kinetics [6]. The resulting models represent multi-dimensional partial integro-differential equations. For efficient numerical solution, a sophisticated approximate moment method was developed and applied to study the effect of host cell heterogeneity in engineered cell lines [2,3,4]. It could be shown that cell-to-cell variability resulting from modification of host-cell factors tends to limit the intended increase in virus yield. In current research efforts, the developed approach was used to investigate cell-to-cell variability in cell lines with multiple gene overexpressions (MGOs). Here, a combinational study was performed to predict the most promising candidates for MGOs thereby reducing potential experimental efforts [5] (see Figure 2).
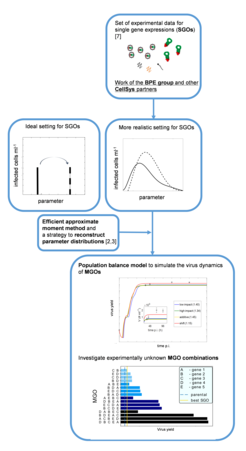
Outlook
Future work will focus on experimental validation of the theoretical predictions for most potential MGO candidates that from the numerical studies. Furthermore, the effects of defective interfering particles (DIPs) [7] on the viral production will be included in the model formulation.
Cooperation Partners
The project was embedded in the CellSys project (grant number 031 6189 A) funded by the German Federal Ministry of Science and Education (Bundesministerium für Bildung und Forschung- BMBF). Cooperation partners in the CellSys project were:
- Prof. Dr.-Ing. Udo Reichl, Max Planck Institute for Dynamics of Complex Technical Systems, Bioprocess Engineering
- Prof. Dr. T. Meyer, Dr. A. Karlas, Max Planck Institute for Infection Biology Berlin, Molecular Biology Unit
- Dr. D. Wirth, Dr. H. Hauser, Helmholtz Centre for Infection Research Brunswick, Molecular Biotechnology
- Dr. V. Sandig, ProBioGen AG, Berlin
- Dr. M. Lesch, Steinbeis gGmbH, Berlin
References
Own Publications:
[1] T. Müller,1 R. Dürr, B. Isken, J.Schulze-Horsel, U. Reichl, A. Kienle. Distributed Modeling of Human Influenza A Virus–Host Cell Interactions During Vaccine Production. Biotechnology and Bioengineering, 110(8), p. 2252-2266, 2013.
[2] R. Dürr, S. Duvigneau, T. Laske, M. Bachmann, A. Kienle. Analyzing the impact of heterogeneity in genetically engineered cell lines for influenza vaccine production using population balance modeling. Proceedings to Foundations of Systems Biology in Engineering – FOSBE 2016, IFAC-PapersOnline, 49, p. 225 – 230, 2016.
[3] R. Dürr. Parameter estimation and method of moments for multi dimensional population balance equations with application to vaccine production processes. PhD thesis, Faculty of electrical engineering and information technology, Otto von Guericke University Magdeburg, 2016
[4] R. Dürr, T. Müller, S. Duvigneau, A. Kienle. An efficient approximate moment method for multi-dimensional population balance models – Application to virus replication in multi-cellular systems. Chemical Engineering Science, 160, p. 321 – 334, 2017.
[5] S. Duvigneau, R. Dürr, T. Laske, M. Bachmann, M. Dostert, U. Reichl, A. Kienle. Mathematical modeling as a tool to improve influenza vaccine production processes. Proceedings to Foundations of Systems Biology in Engineering – FOSBE 2018, IFAC-PapersOnline, 51, p. 1-4, 2018.
Related research articles (BPE group):
[6] F. S. Heldt, T. Frensing, A. Pflugmacher, R. Gröpler, B. Peschel, U. Reichl. Multiscale Modeling of Influenza A Virus Infection Supports the Development of Direct-Acting Antivirals. PLoS Computational Biology. 9(11), 2013.
[7] T. Laske, F. S. Heldt, H. Hoffmann, T. Frensing, U. Reichl. Reprint of Modeling the intracellular replication of influenza A virus in the presence of defective interfering RNAs. Virus Research, 218, p. 86 – 95, 2016.
[8] T. Laske, M. Bachmann, M. Dostert; T. Frensing, A. Karlas, D. Wirth, U. Reichl. Model-based analysis of influenza A virus replication in genetically engineered cell lines elucidates the impact of host cell factors on key kinetic parameters of virus growth. PloS Computational Biology, 2018, under review.

