RNA structure and interactions involved in OP7 DIP genome packaging
Motivation
Conventional defective interfering particles (DIPs) derived from the influenza A virus (IAV) carry a large internal deletion in one of their genomic viral RNA (vRNA). A unique, non-conventional DIP termed “OP7” was discovered recently by our team. It contains 37 point mutations in its segment 7 vRNA, affecting promoter regions, encoded proteins and genome packaging signals. One of the most extensively studied topics is the genome packaging into the progeny virions. This stage of the viral replication cycle is coordinated by complex intersegment vRNA-vRNA interactions, which are intended to ensure specificity of segment incorporation and a proper genomic composition of newly formed virions (Figure 1).
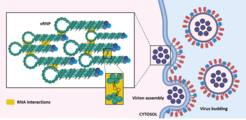
So far, the mechanism of action and the reported phenotypic effects of OP7 are only poorly elucidated, especially at the level of packaging. For example, during STV/OP7 co-infection, the segment composition within the progeny virions is disturbed. More specifically, our studies suggest that OP7 virions show an abnormal vRNA content, where the mutated segment 7 is predominantly packaged into progeny virions. This may be caused by a rearrangement of the vRNA-vRNA interactions enforced by the mutations.
Aim of the project
We utilize RNA structure mapping approaches, binding assays and thorough interaction analysis to gain insight into both intramolecular and intermolecular RNA-RNA interactions, as well as RNA-NP binding. The obtained experimental data facilitate computational structure predictions and will improve their accuracy. The results enable to highlight the differences between WT vRNA and OP7 vRNA interplay and virion assembly. We aim to indicate structural features, which may determine the packaging capacity and interfering potential of the DIP in a co-infection scenario. The acquired knowledge will contribute to a better understanding of OP7 infection dynamics, molecular aspects of co-infection, and further to the development of antiviral particles with higher preventive and therapeutic potential.
