Imaging of Influenza Virus Replication to Optimize Cell Culture-based Vaccine Production
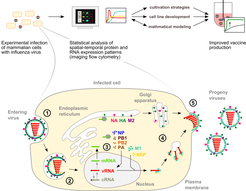
Figure 1: Image analysis of influenza virus replication in animal cells to improve vaccine production.
To identify limitations of influenza vaccine production in animal cell cultures, we investigate influenza virus replication at the cellular and the molecular level. Influenza virus replication occurs in the following regulated steps (1) virus uptake into the cell, (2) virus uncoating and release of the viral genome (vRNA), (3) viral protein synthesis and vRNA replication (4) transport of viral proteins and vRNA to the plasma membrane, and (5) virus assembly and release from the cell surface. The efficiency of influenza virus production largely depends on the capacity of the host cell to support viral replication. Therefore, we analyze the temporal-spatial expression and distribution of host cell and viral components to identify replication patterns and dynamics in highly productive cells. The results will contribute to the design of high-yield cell lines and cultivation strategies for influenza vaccine production.
Abbreviations: cRNA, complementary RNA; HA, hemagglutinin; M1, matrix protein 1; M2, matrix protein 2; mRNA, messenger RNA; NA, neuraminidase; NP, nucleoprotein; NEP, nuclear export protein; PA, RNA polymerase subunit (acidic); PB1, RNA polymerase subunit 1 (basic); PB2, polymerase subunit 2 (basic); vRNA, viral RNA (genome)
© MPI Magdeburg (BPE)
References
Multiscale model of defective interfering particle replication for influenza A virus infection in animal cell culture. PLoS Computational Biology 17, e1009357 (2021)
A Novel Type of Influenza A Virus-Derived Defective Interfering Particle with Nucleotide Substitutions in Its Genome. Journal of Virology 93 (4), 01786-18 (2019)
Influenza virus intracellular replication dynamics, release kinetics, and particle morphology during propagation in MDCK cells. Applied Microbiology and Biotechnology 100 (16), pp. 7181 - 7192 (2016)
